Plannedexperiment Resource¶
Resource URL: http://mytorrentserver/rundb/api/v1/plannedexperiment/
Schema URL: http://mytorrentserver/rundb/api/v1/plannedexperiment/schema/
Perform CRUD operations on plannedexperiment resources and data elements.
Even though plannedExperiment db schema has changed dramatically in TSS 3.6 as part of the “plan data decentralization” (aka PDD) effort. A facade is provided so if you are already familiar with using the plan REST API, changes under the hood are abstracted from the REST API users. However, note that “selectedPlugins” and “barcodedSamples” are JSON fields and their data structures tend to change from release to release.
What has changed in TSS 4.2¶
- The JSON data structure in barcodedSamples has been changed with the following added
- controlSequenceType
- hotSpotRegionBedFile
- nucleotideType
- reference
- targetRegionBedFile
- The JSON data structure in selectedPlugins for IonReporter has been changed with the following added
- NucleotideType
- cancerType
- cellularityPct
- New VariantCaller parameters have been added and some parameters have been obsolete (persisted in selectedPlugins)
- New values for runType, applicationGroup and sampleGrouping have been added to support DNA and Fusions
- Some new attributes intended for internal use only have been added to plannedExperiment.
- We have started enforcing validation during REST API posting for
- barcodeId
- chipType
- flows
- notes
- planName
- project or projects
- runType
- sampleTubeLabel
- sample or sample in barcodedSamples
- sampleGroupingName
- sequencekitname
- templateKitName
- Posting that fails validation will receive an error code.
- Until stringent validation is fully in place during non-GUI REST API posting, please do your due diligence to ensure the data and data format posted are valid.
Moreover, some attributes require “internal” value instead of the “customer-facing” value to be persisted (e.g., sequencekitname, chipType). Please refer to the Comment/Expected Value column more details.
Validation Rules¶
RULE-1: Valid characters: letters, numbers, dashes, underscores, dots
RULE-2: Valid characters: letters, numbers, spaces, dashes, underscores, dots
RULE-3: Invalid leading characters: dashes, underscores, dots
Field Notes¶
| Attribute Name | Required/Optional/Nullable | Data type | Default value | Valid values | Example | Comment/Expected Value |
|---|---|---|---|---|---|---|
| adapter | Opt/Nullable | varchar(256) | Not really being used | |||
| applicationGroupDisplayedNamne | Opt/Nullable | DNA, DNA and Fusions, Metagenomics, RNA, Typing | ||||
| autoAnalyze | Boolean | TRUE | ||||
| autoName | Opt/Nullable | varchar(512) | Not really being used | |||
| barcodeId | Opt/Nullable | varchar(128) | IonSet1 | rundb_dnabarcode.name | ||
| barcodedSamples | Opt/Nullable | json | refer to example below | |||
| base_recalibrate | Opt | Boolean | whether to recalibrate signal measurements for homo-polymers | |||
| bedfile | Opt/Nullable | varchar(1024) | /results/uploads/BED/71/hg19/unmerged/detail/CFTRexon.20131001.designed.bed | target region BED file rundb_content.path | ||
| chipBarcode | Opt/Nullable | varchar(64) | ||||
| chipType | Opt | varchar(32) | 318v2 | rundb_chip.name Even though REST API posting will allow you to create a plan without specifying the chipType, TS UI will require chipType to be specified. | ||
| controlSequencekitname | Opt/Nullable | varchar(512) | rundb_kitInfo.name | |||
| cycles | Opt/Nullable | int | ||||
| date | Opt/Nullable | DateTimeField | ||||
| expName | Opt | varchar(128) | Do not set the value manually. Crawler will set it during explog processing | |||
| flows | Req | int | 0 | 500 | ||
| flowsInOrder | Opt/Nullable | varchar(512) | Do not set the value manually | |||
| forward3primeadapter | Req | varchar(512) | ATCACCGACTGCCCATAGAGAGGCTGAGAC | |||
| id | Opt | int | Do not set this value unless you are updating a plan | |||
| irworkflow | Opt | varchar(1024) | TSS 2.4/IonReporter-related; no longer being used | |||
| isDuplicateReads | Opt | Boolean | Whether to filter out PCR duplicates | |||
| isFavorite | Opt | Boolean | FALSE | |||
| isPlanGroup | Opt | Boolean | FALSE | |||
| isReusable | Opt | Boolean | FALSE | |||
| isReverseRun | Req | Boolean | FALSE | True,False | ||
| isSystem | Opt | Boolean | FALSE | |||
| isSystemDefault | Opt | Boolean | FALSE | |||
| libkit | Opt/Nullable | varchar(512) | Ion Xpress Plus Fragment Library Kit | rundb_kitInfo.name | ||
| library | Opt/Nullable | varchar(512) | hg19 | rundb_referencegenome.short_name | ||
| libraryKey | Req | varchar(64) | TCAG | |||
| librarykitname | Opt/Nullable | varchar(512) | Ion AmpliSeq 2.0 Library Kit | rundb_kitInfo.name | ||
| metaData | Opt | json | ||||
| notes | Opt/Nullable | varchar(1024) | see RULE-2 | |||
| pairedEndLibraryAdapterName | Opt/Nullable | varchar(512) | Since paired-end sequencing has been dis-continued, do not use. | |||
| parentPlan | Opt/Nullable | FK | Currently used for paired-end plans only. Since PE plans have been dis-continued, do not use. | |||
| planDisplayedName | varchar(512) | demo plan | see RULE-2 REST API posting does not support this attribute yet. Use planName instead. | |||
| planExecuted | Opt | Boolean | FALSE | True,False | ||
| planExecutedDate | Opt/Nullable | DateTimeField | ||||
| planGUID | Opt/Nullable | varchar( 512) | Do not set a value manually during plan creation | |||
| planName | varchar(512) | demo_plan | see RULE-1 | |||
| planPGM | Opt/Nullable | varchar(128) | Not being used | |||
| platform | Opt | varchar(128) | “” | “”, PGM, PROTON | ||
| planShortID | Opt/Nullable | Do not set a value manually during plan creation | ||||
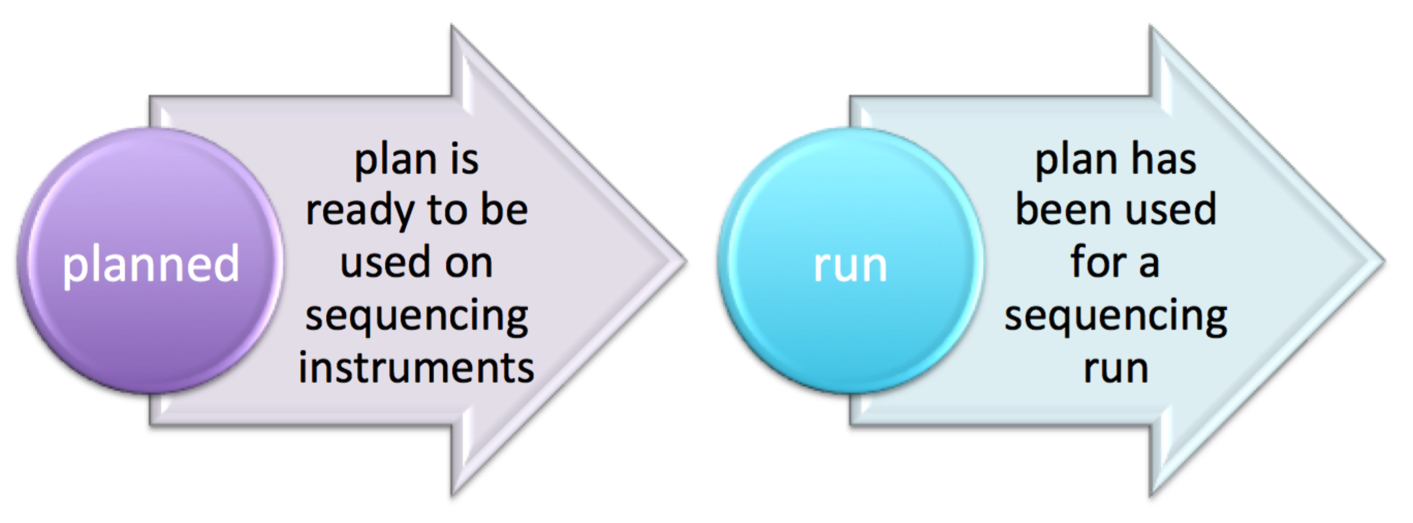
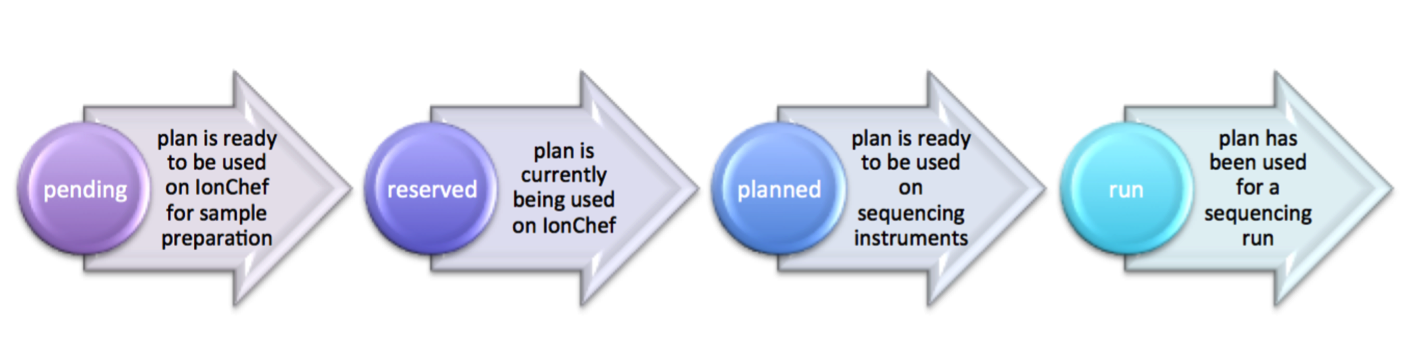
| planStatus | varchar(512) | planned | “”, pending, reserved, planned, run | see planStatus state diagrams below For OneTouch & IonChef | ||
| preAnalysis | Opt | Boolean | ||||
| projects | Opt | varchar(64) for each project name | [“project1”,”project2”] | see RULE-1 a list of comma separated project names | ||
| realign | Opt | Boolean | whether to run an optional analysis step to adjust the alignment, primarily in the CIGAR string | |||
| regionfile | Opt/Nullable | varchar(1024) | /results/uploads/BED/71/hg19/unmerged/detail/CFTRexon.20131125.hotspots.bed | hotspot region BED file | ||
| reverse_primer | Opt/Nullable | varchar(128) | ||||
| runMode | Opt | varchar(64) | “”,”single”, | single | ||
| runType | Req | varchar(512) | GENS | “AMPS”, “AMPS_DNA_RNA”, “AMPS_EXOME”, “AMPS_RNA”, “GENS”, “RNA”, “TAR”, “WGNM”, “TARS_16S” | rundb_runtype.runType | |
| runName | Opt/Nullable | varchar(255) | Not being used | |||
| sample | Required for plan | varchar(127) | demo_sample | see RULE-1, RULE-3 | ||
| sampleDisplayedName | Opt/Nullable | varchar(127) | demo sample | see RULE-2, RULE-3 REST API posting does not support this attribute yet. Use sample instead. | ||
| sampleGroupingName | Opt/Nullable | DNA_RNA, Other, Sample_Control, Self, Tumor_Normal | Self | |||
| samplePrepKitName | Opt/Nullable | varchar(512) | Ion TargetSeq(tm) Custom Enrichment Kit (100kb-500kb) | rundb_kitInfo.name | ||
| sampleTubeLabel | Opt/Nullable | varchar(512) | X12450aab | The barcode on the tube that contains the sample genetic material for sequencing | ||
| selectedPlugins | Opt/Nullable | json | refer to example below | Since plugin configuration parameters are stored with the selected plugins, it can get complicated fast. It is not advised to manually post the selectedPlugins json blob. | ||
| seqKitBarcode | Opt/Nullable | varchar(64) | Not really being used | |||
| sequencekitname | Recommend to set | varchar(512) | IonPGM200Kit-v2 | rundb_kitInfo.name | ||
| storageHost | Opt/Nullable | varchar(128) | ||||
| storage_options | Opt | varchar(200) | A | “KI”,”A”,”D” | ||
| templatingKitName | Opt/Nullable | varchar(512) | Ion PGM Template OT2 200 Kit | for either OneTouch or IonChef rundb_kitInfo.name | ||
| usePostBeadfind | Opt | Boolean | ||||
| usePreBeadfind | Opt | Boolean | TRUE | |||
| username | Opt/Nullable | varchar(128) | ionuser | the user currently logs in to Torrent Browser for this GUI-based plan creation. For REST API posting, this is just treated as freeform text auth_user.username |
barcodedSamples JSON Examples¶
Generic sequencing plan¶
"barcodedSamples": {
"s 1": {
"barcodeSampleInfo": {
"IonSet1_16": {
"controlSequenceType": "",
"description": "desc 1",
"externalId": "accession 101",
"hotSpotRegionBedFile": "/results/uploads/BED/19/hg19/unmerged/detail/4477685_CCP_hotspots_20121225.bed",
"nucleotideType": "DNA",
"reference": "hg19",
"targetRegionBedFile": "/results/uploads/BED/19/hg19/unmerged/detail/4477685_CCP_designed.bed"
}
},
"barcodes": [
"IonSet1_16"
]
},
"s 2": {
"barcodeSampleInfo": {
"IonSet1_12": {
"controlSequenceType": "",
"description": "desc 2",
"externalId": "accession 80",
"hotSpotRegionBedFile": "/results/uploads/BED/19/hg19/unmerged/detail/4477685_CCP_hotspots_20121225.bed",
"nucleotideType": "DNA",
"reference": "hg19",
"targetRegionBedFile": "/results/uploads/BED/19/hg19/unmerged/detail/4477685_CCP_designed.bed"
}
},
"barcodes": [
"IonSet1_12"
]
},
"s 3": {
"barcodeSampleInfo": {
"IonSet1_15": {
"controlSequenceType": "",
"description": "desc 3",
"externalId": "accession 280",
"hotSpotRegionBedFile": "/results/uploads/BED/19/hg19/unmerged/detail/4477685_CCP_hotspots_20121225.bed",
"nucleotideType": "DNA",
"reference": "hg19",
"targetRegionBedFile": "/results/uploads/BED/19/hg19/unmerged/detail/4477685_CCP_designed.bed"
}
},
"barcodes": [
"IonSet1_15"
]
}
},
Onconet DNA plan¶
"barcodedSamples": {
"example 1": {
"barcodeSampleInfo": {
"IonXpress_010": {
"controlSequenceType": "",
"description": "example here",
"externalId": "id 1",
"hotSpotRegionBedFile": "/results/uploads/BED/22/hg19/unmerged/detail/ColonLung.20131001.hotspots.bed",
"nucleotideType": "DNA",
"reference": "hg19",
"targetRegionBedFile": "/results/uploads/BED/22/hg19/unmerged/detail/ColonLung.20131001.designed.bed"
}
},
"barcodes": [
"IonXpress_010"
]
},
"example 2": {
"barcodeSampleInfo": {
"IonXpress_005": {
"controlSequenceType": "",
"description": "another example here",
"externalId": "id 2",
"hotSpotRegionBedFile": "/results/uploads/BED/22/hg19/unmerged/detail/ColonLung.20131001.hotspots.bed",
"nucleotideType": "DNA",
"reference": "hg19",
"targetRegionBedFile": "/results/uploads/BED/22/hg19/unmerged/detail/ColonLung.20131001.designed.bed"
}
},
"barcodes": [
"IonXpress_005"
]
}
},
Onconet DNA and Fusions plan¶
"barcodedSamples": {
"s 1": {
"barcodeSampleInfo": {
"IonXpress_001": {
"controlSequenceType": "",
"description": "description here",
"externalId": "ext 1",
"hotSpotRegionBedFile": "/results/uploads/BED/22/hg19/unmerged/detail/ColonLung.20131001.hotspots.bed",
"nucleotideType": "DNA",
"reference": "hg19",
"targetRegionBedFile": "/results/uploads/BED/22/hg19/unmerged/detail/ColonLung.20131001.designed.bed"
},
"IonXpress_002": {
"controlSequenceType": "",
"description": "description here",
"externalId": "ext 1",
"hotSpotRegionBedFile": "",
"nucleotideType": "RNA",
"reference": "hg19_rna",
"targetRegionBedFile": ""
}
},
"barcodes": [
"IonXpress_001",
"IonXpress_002"
]
}
}
selectedPlugins JSON Examples¶
IonReporterUploader, coverageAnalysis, sampleId and variantCaller¶
"selectedPlugins": {
"IonReporterUploader": {
"features": [
"export"
],
"id": 700,
"name": "IonReporterUploader",
"userInput": {
"accountId": "1234567890abcde",
"accountName": " demo IonReporter (Version: 4.2 | User: Ion User | Org: IR Org)",
"userInputInfo": [{
"ApplicationType": "Low-Coverage Whole Genome Sequencing",
"Gender": "Female",
"NucleotideType": "DNA",
"Relation": "Self",
"RelationRole": "Self",
"Workflow": "Test_WK_1",
"barcodeId": "IonXpress_010",
"cancerType": "Breast Cancer",
"cellularityPct": "23",
"sample": "example 1",
"sampleDescription": "example here",
"sampleExternalId": "id 1",
"sampleName": "example_1",
"setid": "1__4c310e03-d188-4702-b82a-f9043bc04350"
}, {
"ApplicationType": "Low-Coverage Whole Genome Sequencing",
"Gender": "Male",
"NucleotideType": "DNA",
"Relation": "",
"RelationRole": "Self",
"Workflow": "Test_WK_1",
"barcodeId": "IonXpress_005",
"cancerType": "Liver Cancer",
"cellularityPct": "27",
"sample": "example 2",
"sampleDescription": "another example here",
"sampleExternalId": "id 2",
"sampleName": "example_2",
"setid": "2__4c310e03-d188-4702-b82a-f9043bc04350"
}]
},
"version": "4.2-r88003"
},
"coverageAnalysis": {
"features": [],
"id": 696,
"name": "coverageAnalysis",
"userInput": "",
"version": "4.2-r87890"
},
"sampleID": {
"features": [],
"id": 701,
"name": "sampleID",
"userInput": "",
"version": "4.2-r87942"
},
"variantCaller": {
"features": [],
"id": 699,
"name": "variantCaller",
"userInput": {
"freebayes": {
"allow_complex": "0",
"allow_indels": "1",
"allow_mnps": "0",
"allow_snps": "1",
"gen_min_alt_allele_freq": "0.03",
"gen_min_coverage": "6",
"gen_min_indel_alt_allele_freq": "0.1",
"min_base_qv": "2",
"min_mapping_qv": "4",
"read_max_mismatch_fraction": "1.0",
"read_mismatch_limit": "10"
},
"long_indel_assembler": {
"kmer_len": "19",
"max_hp_length": "8",
"min_indel_size": "4",
"min_var_count": "5",
"min_var_freq": "0.15",
"relative_strand_bias": "0.8",
"short_suffix_match": "5"
},
"meta": {
"built_in": true,
"compatibility": {
"chip": [
"pgm",
"proton_p1"
],
"library": [
"ampliseq"
],
"panel": "/rundb/api/v1/contentupload/22/"
},
"configuration": "",
"librarytype": "ampliseq",
"name": "Panel-optimized - Colon and Lung Panel - 10/7/2013",
"repository_id": "",
"tooltip": "Panel-optimized parameters from AmpliSeq.com",
"trimreads": true,
"ts_version": "4.0",
"tvcargs": "tvc",
"user_selections": {
"chip": "pgm",
"frequency": "germline",
"library": "ampliseq",
"panel": "/rundb/api/v1/contentupload/22/"
}
},
"torrent_variant_caller": {
"data_quality_stringency": "6.5",
"downsample_to_coverage": "10000",
"filter_deletion_predictions": "0.2",
"filter_insertion_predictions": "0.2",
"filter_unusual_predictions": "0.3",
"heavy_tailed": "3",
"hotspot_beta_bias": "100.0",
"hotspot_min_allele_freq": "0.01",
"hotspot_min_cov_each_strand": "2",
"hotspot_min_coverage": "6",
"hotspot_min_variant_score": "6",
"hotspot_strand_bias": "0.95",
"hp_max_length": "8",
"indel_beta_bias": "10.0",
"indel_min_allele_freq": "0.05",
"indel_min_cov_each_strand": "2",
"indel_min_coverage": "15",
"indel_min_variant_score": "6",
"indel_strand_bias": "0.9",
"outlier_probability": "0.01",
"prediction_precision": "1.0",
"snp_beta_bias": "100.0",
"snp_min_allele_freq": "0.02",
"snp_min_cov_each_strand": "0",
"snp_min_coverage": "6",
"snp_min_variant_score": "6",
"snp_strand_bias": "0.95"
}
bbb
},
"version": "4.2-r87667"
}
},
"seqKitBarcode": null,
"sequencekitname": "IonPGM200Kit-v2",
"storageHost": null,
"storage_options": "A",
"templatingKitBarcode": null,
"templatingKitName": "Ion PGM Template OT2 200 Kit",
"tfKey": "ATCG",
"thumbnailalignmentargs": "",
"thumbnailanalysisargs": "",
"thumbnailbasecallerargs": "",
"thumbnailbeadfindargs": "",
"thumbnailcalibrateargs": "",
"usePostBeadfind": true,
"usePreBeadfind": true,
"username": "ionadmin",
"variantfrequency": ""
},
IonReporterUploader selected for a Onconet DNA and Fusions plan¶
"selectedPlugins": {
"IonReporterUploader": {
"features": [
"export"
],
"id": 700,
"name": "IonReporterUploader",
"userInput": {
"accountId": "1234567890abcde ",
"accountName": "demo IonReporter (Version: 4.2 | User: Ion User | Org: IR Org)",
"userInputInfo": [{
"ApplicationType": "Oncomine_DNA_RNA_Fusion",
"Gender": "Male",
"NucleotideType": "DNA",
"Relation": "DNA_RNA",
"RelationRole": "Self",
"Workflow": "AmpliSeq Colon Lung v2 with RNA Lung Fusion single sample",
"barcodeId": "IonXpress_001",
"cancerType": "Colorectal Cancer",
"cellularityPct": "17",
"sample": "s 1",
"sampleDescription": "description here",
"sampleExternalId": "ext 1",
"sampleName": "s_1",
"setid": "1__381a5a84-5af0-40ff-84c1-b31720fea6ca"
}, {
"ApplicationType": "Oncomine_DNA_RNA_Fusion",
"Gender": "Male",
"NucleotideType": "RNA",
"Relation": "DNA_RNA",
"RelationRole": "Self",
"Workflow": "AmpliSeq Colon Lung v2 with RNA Lung Fusion single sample",
"barcodeId": "IonXpress_002",
"cancerType": "Colorectal Cancer",
"cellularityPct": "17",
"sample": "s 1",
"sampleDescription": "description here",
"sampleExternalId": "ext 1",
"sampleName": "s_1",
"setid": "1__381a5a84-5af0-40ff-84c1-b31720fea6ca"
}]
},
"version": "4.2-r88003"
}
},
Creating a plan¶
Non-barcoded PGM¶
Post a non-barcoded Target Sequencing PGM plan and to associate results with 2 projects with sampleGrouping and applicationGroup specified:
{
"autoAnalyze": "true",
"usePreBeadfind": "true",
"usePostBeadfind": "true",
"reverselibrarykey": "",
"reverse3primeadapter": "",
"libraryKey": "TCAG",
"forw ard3primeadapter": "ATCACCGACTGCCCATAGAGAGGCTGAGAC",
"flows": 500,
"library": "hg19",
"bedfile": "/results/uploads/BED/71/hg19/unmerged/detail/CFTRexon.20131001.designed.bed",
"regionfile": "/results/uploads/BED/71/hg19/unmerged/detail/CFTRexon.20131125.hotspots.bed",
"planName": "DEMO-TS4_2_x-REST- API_TARS_plan1",
"sample": "my_sample",
"notes": "this is a REST test plan",
"username": "ionuser",
"preAnalysis": "on",
"isReverseRun": false,
"isPlanGroup": false,
"runMode": "single",
"runType": "TARS",
"chipType": "318v2",
"sequencekitname": "IonPGM200Kit",
"librarykitname": "Ion Xpress Plus Fragment Library Kit",
"templatingKitName": "Ion PGM Template OT2 200 Kit",
"samplePrepKitName": "Ion TargetSeq(tm) Custom Enrichment Kit (100kb-500kb)",
"projects": ["myProject1", "myProject2"],
"sampleGroupingName": "Self",
"applicationGroupDisplayedName": "DNA"
}
Non-Barcoded PI¶
Post a non-barcoded Target Sequencing Proton plan with PI chip, with sample tube label, chip barcode and the QC thresholds specified:
{
"autoAnalyze": "true",
"usePreBeadfind": "true",
"usePostBeadfind": "true",
"reverselibrarykey": "",
"reverse3primeadapter": "",
"libraryKey": "TCAG",
"forward3primeadapter": "ATCACCGACTGCCCATAGAGAGGCTGAGAC",
"flows": 440,
"library": "hg19",
"bedfile": "/results/uploads/BED/14/hg19/unmerged/detail/BRCA1_2.20131001.designed.bed",
"regionfile": "/results/uploads/BED/14/hg19/unmerged/detail/BRCA1_2.20131001.hotspots.bed",
"planName": "DEMO-TS4_2_x-REST-API_TARS_Proton_plan2",
"sample": "my_sample",
"notes": "here are my notes",
"username": "ionuser",
"preAnalysis": "on",
"isReverseRun": false,
"isPlanGroup": false,
"runMode": "single",
"runType": "TARS",
"chipType": "P1.1.17",
"sequencekitname": "ProtonI200Kit-v3",
"librarykitname": "Ion Xpress Plus Fragment Library Kit",
"templatingKitName": "Ion PI Template OT2 200 Kit v3",
"samplePrepKitName": "Ion TargetSeq(tm) Exome Kit (4 rxn)",
"projects": ["myProject1"],
"sampleTubeLabel": "abcX254",
"chipBarcode": "AA02314571",
"Bead Loading (%)": 33,
"Key Signal (1-100)": 35,
"Usable Sequence (%)": 37
}
Barcoded RNA PGM¶
Post a barcoded RNA Sequencing PGM plan:
{
"autoAnalyze": "true",
"usePreBeadfind": "true",
"usePostBeadfind": "true",
"reverselibrarykey": "",
"reverse3primeadapter": "",
"libraryKey": "TCAG",
"forward3primeadapter": "ATCACCGACTGCCCATAGAGAGGCTGAGAC",
"flows": 160,
"library": "hg19_rna",
"planName": "DEMO-TS4_2_x-REST- API_barcoded_RNA_plan3",
"notes": "test notes here ",
"username": "ionuser",
"preAnalysis": "on",
"isReverseRun": false,
"isPlanGroup": false,
"runMode": "single",
"runType": "RNA",
"chipType": "318v2",
"sequencekitname": "IonPGM200Kit-v2",
"librarykitname": "Ion Total RNA Seq Kit v2",
"templatingKitName": "Ion PGM Template OT2 200 Kit",
"samplePrepKitName": "",
"projects": ["myProject1", "myProject2"],
"barcodedSamples": "{'demo sample 1':{'barcodeSampleInfo':{'IonXpressRNA_003':{'controlSequenceType' : 'ERCC Mix 1', 'externalId':'x 1','description':'description here', 'hotSpotRegionBedFile':'', 'nucleotideType': 'RNA', 'reference': 'hg19_rna', 'targetRegionBedFile': ''}},'barcodes':['IonXpressRNA_003']},'demo sample 2':{'barcodeSampleInfo':{'IonXpressRNA_004':{'controlSequenceType' : 'ERCC Mix 2', 'externalId':'x 2','description':'description there', 'hotSpotRegionBedFile':'', 'nucleotideType': 'RNA', 'reference': 'hg19_rna', 'targetRegionBedFile': ''}},'barcodes':['IonXpressRNA_004']}}",
"applicationGroupDisplayedName": "RNA",
"barcodeId": "IonXpressRNA",
"sampleTubeLabel": "2554abc",
"Bead Loading (%)": 30,
"Key Signal (1-100)": 30,
"Usable Sequence (%)": 30
}
Using POST to update a plan¶
If you are to update a plan via REST API, please perform a GET first so you’ll have all the internally created values for the plan to perform the update with a POST.
To update with a POST, just include “id”: <plan PK> in your data packet (e.g., “id”:1234)
About using PUT or PATCH to update a plan¶
Update a plan for its chipBarcode value
http://<hostname>/rundb/api/v1/plannedexperiment/<plan pk>/?format=json
{
"chipBarcode": "AA323323"
}
Fields table¶
| field | help text | default | nullable | readonly | blank | unique | type |
|---|---|---|---|---|---|---|---|
| planDisplayedName | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| autoAnalyze | Boolean data. Ex: True | n/a | false | false | false | false | boolean |
| templatingKitBarcode | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| preAnalysis | Boolean data. Ex: True | false | false | true | false | boolean | |
| chefStatus | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| applicationGroup | A single related resource. Can be either a URI or set of nested resource data. | n/a | true | false | true | false | related |
| libkit | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| platform | Unicode string data. Ex: “Hello World” | n/a | true | true | true | false | string |
| categories | Unicode string data. Ex: “Hello World” | true | false | false | false | string | |
| planPGM | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| sampleSet_planTotal | Integer data. Ex: 2673 | 0 | false | false | false | false | integer |
| projects | Many related resources. Can be either a list of URIs or list of individually nested resource data. | n/a | true | false | true | false | related |
| notes | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| sequencekitname | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| storageHost | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| expName | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| cycles | Integer data. Ex: 2673 | n/a | true | false | false | false | integer |
| isReverseRun | Boolean data. Ex: True | false | false | false | true | false | boolean |
| storage_options | Unicode string data. Ex: “Hello World” | A | false | false | false | false | string |
| chipType | Unicode string data. Ex: “Hello World” | false | false | false | false | string | |
| chefProgress | Floating point numeric data. Ex: 26.73 | 0.0 | false | false | true | false | float |
| library | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| reverselibrarykey | Unicode string data. Ex: “Hello World” | false | true | false | false | string | |
| sampleTubeLabel | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| seqKitBarcode | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| barcodeId | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| chefLogPath | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| isPlanGroup | Boolean data. Ex: True | false | false | false | true | false | boolean |
| realign | Boolean data. Ex: True | n/a | false | false | false | false | boolean |
| sampleGroupingName | Unicode string data. Ex: “Hello World” | n/a | true | true | true | false | string |
| experiment | A single related resource. Can be either a URI or set of nested resource data. | n/a | true | false | true | false | related |
| bedfile | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| isReusable | Boolean data. Ex: True | false | false | false | true | false | boolean |
| isDuplicateReads | Boolean data. Ex: True | n/a | false | false | false | false | boolean |
| librarykitname | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| adapter | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| tfKey | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| parentPlan | Unicode string data. Ex: “Hello World” | None | false | false | true | false | string |
| forward3primeadapter | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| chefLastUpdate | A date & time as a string. Ex: “2010-11-10T03:07:43” | n/a | true | false | false | false | datetime |
| samplePrepKitName | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| applicationGroupDisplayedName | Unicode string data. Ex: “Hello World” | n/a | true | true | true | false | string |
| metaData | Unicode string data. Ex: “Hello World” | {} | false | false | true | false | string |
| sampleSet_uid | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| isFavorite | Boolean data. Ex: True | false | false | false | true | false | boolean |
| sampleSet_planIndex | Integer data. Ex: 2673 | 0 | false | false | false | false | integer |
| qcValues | Many related resources. Can be either a list of URIs or list of individually nested resource data. | n/a | true | false | true | false | related |
| planStatus | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| templatingKitName | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| runType | Unicode string data. Ex: “Hello World” | GENS | false | false | false | false | string |
| username | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| planName | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| sampleDisplayedName | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| controlSequencekitname | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| chefMessage | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| childPlans | A list of data. Ex: [‘abc’, 26.73, 8] | [] | false | false | false | false | list |
| pairedEndLibraryAdapterName | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| runMode | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| irworkflow | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| planExecuted | Boolean data. Ex: True | false | false | false | true | false | boolean |
| project | Unicode string data. Ex: “Hello World” | n/a | false | true | true | false | string |
| usePostBeadfind | Boolean data. Ex: True | false | false | true | false | boolean | |
| runname | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| planGUID | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| planShortID | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| sampleSetGroupType | Unicode string data. Ex: “Hello World” | n/a | true | true | true | false | string |
| sample | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| planExecutedDate | A date & time as a string. Ex: “2010-11-10T03:07:43” | n/a | true | false | false | false | datetime |
| reverse_primer | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| id | Integer data. Ex: 2673 | false | false | true | true | integer | |
| barcodedSamples | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| regionfile | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| selectedPlugins | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| sampleSet | A single related resource. Can be either a URI or set of nested resource data. | n/a | true | false | true | false | related |
| isSystemDefault | Boolean data. Ex: True | false | false | false | true | false | boolean |
| autoName | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| libraryKey | Unicode string data. Ex: “Hello World” | false | false | true | false | string | |
| flows | Integer data. Ex: 2673 | 0 | false | false | false | false | integer |
| date | A date & time as a string. Ex: “2010-11-10T03:07:43” | n/a | true | false | false | false | datetime |
| isSystem | Boolean data. Ex: True | false | false | false | true | false | boolean |
| variantfrequency | Unicode string data. Ex: “Hello World” | false | true | false | false | string | |
| sampleSetDisplayedName | Unicode string data. Ex: “Hello World” | n/a | true | true | true | false | string |
| flowsInOrder | Unicode string data. Ex: “Hello World” | true | false | true | false | string | |
| sampleGrouping | A single related resource. Can be either a URI or set of nested resource data. | n/a | true | false | true | false | related |
| base_recalibrate | Boolean data. Ex: True | n/a | false | false | false | false | boolean |
| chipBarcode | Unicode string data. Ex: “Hello World” | n/a | true | false | false | false | string |
| usePreBeadfind | Boolean data. Ex: True | false | false | true | false | boolean | |
| resource_uri | Unicode string data. Ex: “Hello World” | n/a | false | true | false | false | string |
| reverse3primeadapter | Unicode string data. Ex: “Hello World” | false | true | false | false | string |
Example request¶
Request URL: http://mytorrentserver/rundb/api/v1/plannedexperiment/?format=json&limit=1
Python example¶
import requests
ts_api_request = requests.get("http://mytorrentserver/rundb/api/v1/plannedexperiment/", params={"format": "json", "limit": 1})
ts_api_response = ts_api_request.json()
plannedexperiments = ts_api_response["objects"]
for plannedexperiment in plannedexperiments:
print plannedexperiment
Torrent Server response¶
{
"meta": {
"previous": null,
"total_count": 18369,
"offset": 0,
"limit": 1,
"next": "/rundb/api/v1/plannedexperiment/?offset=1&limit=1&format=json"
},
"objects": [
{
"planDisplayedName": "CopyOfSystemDefault_R_2014_06_27_14_19_01_user_P19-606",
"autoAnalyze": true,
"templatingKitBarcode": null,
"preAnalysis": true,
"chefStatus": "",
"applicationGroup": "/rundb/api/v1/applicationgroup/1/",
"libkit": null,
"platform": "PROTON",
"categories": "",
"planPGM": null,
"prebasecallerargs": "BaseCaller --barcode-filter 0.01 --barcode-filter-minreads 10 --keypass-filter on --phasing-residual-filter=2.0 --num-unfiltered 1000 --calibration-training=100000 --flow-signals-type scaled-residual",
"alignmentargs": "stage1 map4",
"thumbnailbasecallerargs": "BaseCaller --barcode-filter 0.01 --barcode-filter-minreads 10 --keypass-filter on --phasing-residual-filter=2.0 --num-unfiltered 100000",
"sampleSet_planTotal": 0,
"projects": [],
"notes": "",
"sequencekitname": "ProtonI200Kit-v3",
"storageHost": null,
"expName": "R_2014_06_27_14_19_01_user_P19-606",
"cycles": null,
"isReverseRun": false,
"storage_options": "A",
"thumbnailalignmentargs": "stage1 map4",
"chipType": "P1.0.20",
"chefProgress": 0.0,
"library": "hg19",
"reverselibrarykey": "",
"sampleTubeLabel": null,
"seqKitBarcode": null,
"barcodeId": "",
"chefLogPath": null,
"isPlanGroup": false,
"realign": false,
"sampleGroupingName": "",
"experiment": "/rundb/api/v1/experiment/23975/",
"bedfile": "",
"isReusable": false,
"isDuplicateReads": false,
"thumbnailbeadfindargs": "justBeadFind --beadfind-minlivesnr 3 --region-size=50,50 --beadfind-thumbnail 1 --bkg-use-proton-well-correction false --bkg-exp-tail-fit false --bkg-pca-dark-matter true",
"librarykitname": "Ion Xpress Plus Fragment Library Kit",
"adapter": null,
"basecallerargs": "BaseCaller --barcode-filter 0.01 --barcode-filter-minreads 10 --keypass-filter on --phasing-residual-filter=2.0 --num-unfiltered 1000 --barcode-filter-postpone 1",
"tfKey": "ATCG",
"parentPlan": null,
"forward3primeadapter": "ATCACCGACTGCCCATAGAGAGGCTGAGAC",
"planStatus": "run",
"chefLastUpdate": null,
"samplePrepKitName": null,
"applicationGroupDisplayedName": "DNA",
"metaData": {},
"sampleSet_uid": null,
"isFavorite": false,
"sampleSet_planIndex": 0,
"qcValues": [
{
"threshold": 30,
"plannedExperiment": "/rundb/api/v1/plannedexperiment/102212/",
"id": 266571,
"qcType": {
"description": "",
"minThreshold": 0,
"maxThreshold": 100,
"defaultThreshold": 30,
"qcName": "Bead Loading (%)",
"id": 1,
"resource_uri": "/rundb/api/v1/qctype/1/"
},
"resource_uri": "/rundb/api/v1/plannedexperimentqc/266571/"
},
{
"threshold": 30,
"plannedExperiment": "/rundb/api/v1/plannedexperiment/102212/",
"id": 266570,
"qcType": {
"description": "",
"minThreshold": 1,
"maxThreshold": 100,
"defaultThreshold": 30,
"qcName": "Key Signal (1-100)",
"id": 2,
"resource_uri": "/rundb/api/v1/qctype/2/"
},
"resource_uri": "/rundb/api/v1/plannedexperimentqc/266570/"
},
{
"threshold": 30,
"plannedExperiment": "/rundb/api/v1/plannedexperiment/102212/",
"id": 266569,
"qcType": {
"description": "",
"minThreshold": 0,
"maxThreshold": 100,
"defaultThreshold": 30,
"qcName": "Usable Sequence (%)",
"id": 3,
"resource_uri": "/rundb/api/v1/qctype/3/"
},
"resource_uri": "/rundb/api/v1/plannedexperimentqc/266569/"
}
],
"analysisargs": "Analysis --from-beadfind --clonal-filter-bkgmodel true --region-size=80,112 --bkg-bfmask-update false --gpuWorkLoad 1 --total-timeout 600 --bkg-use-proton-well-correction false --bkg-exp-tail-fit false --bkg-pca-dark-matter true",
"thumbnailcalibrateargs": "calibrate --skipDroop",
"templatingKitName": "Ion PI Template OT2 200 Kit v3",
"runType": "GENS",
"username": null,
"planName": "CopyOfSystemDefault_R_2014_06_27_14_19_01_user_P19-606",
"sampleDisplayedName": "",
"prethumbnailbasecallerargs": "BaseCaller --barcode-filter 0.01 --barcode-filter-minreads 10 --keypass-filter on --phasing-residual-filter=2.0 --num-unfiltered 100000 --calibration-training=100000 --flow-signals-type scaled-residual",
"controlSequencekitname": null,
"chefMessage": "",
"childPlans": [],
"pairedEndLibraryAdapterName": null,
"runMode": "single",
"irworkflow": "",
"planExecuted": true,
"project": "",
"usePostBeadfind": false,
"runname": null,
"planGUID": "80384c15-e0e2-4909-9d4c-950731bf1cf4",
"planShortID": "G5B50",
"sampleSetGroupType": null,
"sample": "",
"planExecutedDate": null,
"reverse_primer": null,
"id": 102212,
"barcodedSamples": {},
"regionfile": "",
"selectedPlugins": {},
"beadfindargs": "justBeadFind --beadfind-minlivesnr 3 --region-size=80,112 --total-timeout 600 --bkg-use-proton-well-correction false --bkg-exp-tail-fit false --bkg-pca-dark-matter true",
"sampleSet": null,
"isSystemDefault": false,
"autoName": null,
"libraryKey": "TCAG",
"flows": 260,
"thumbnailanalysisargs": "Analysis --from-beadfind --clonal-filter-bkgmodel true --region-size=50,50 --bkg-bfmask-update false --gpuWorkLoad 1 --bkg-debug-param 1 --beadfind-thumbnail 1 --bkg-use-proton-well-correction false --bkg-exp-tail-fit false --bkg-pca-dark-matter true",
"date": "2014-06-27T23:36:06.000229+00:00",
"isSystem": false,
"variantfrequency": "",
"sampleSetDisplayedName": "",
"calibrateargs": "calibrate --skipDroop",
"flowsInOrder": "TACGTACGTCTGAGCATCGATCGATGTACAGC",
"sampleGrouping": null,
"base_recalibrate": true,
"chipBarcode": null,
"usePreBeadfind": true,
"resource_uri": "/rundb/api/v1/plannedexperiment/102212/",
"reverse3primeadapter": ""
}
]
}
Allowed HTTP methods¶
- get
- post
- put
- delete
- patch